Although many covalent drugs were serendipitously discovered historically, covalent inhibitors were in general discouraged due to the concerns over their interference with biological assays and potential lack of selectivity. Over the past decades, rational design of covalent inhibitors has gained traction and garnered increased interest, particularly due to their stronger potency, prolonged target engagement, increased selectivity, and effectiveness to some drug-resistance mutants to reversible inhibitors.
Covalent Ligand Discovery Approaches
Reversible ligands can be rationally designed to covalent inhibitors by incorporating reactive warheads to proper positions. The reversible portion offers binding affinity (Ki) while the electrophile reacts with specific amino acids (cysteine, serine, lysine, etc.) and offers covalent interaction (Kinact). The discovery of EGFR (Epidermal Growth Factor Receptor) and BTK (Bruton’s Tyrosine Kinase) covalent inhibitors shared this strategy with ligand discovered first and Michael acceptor electrophiles incorporated later (Figure 1).
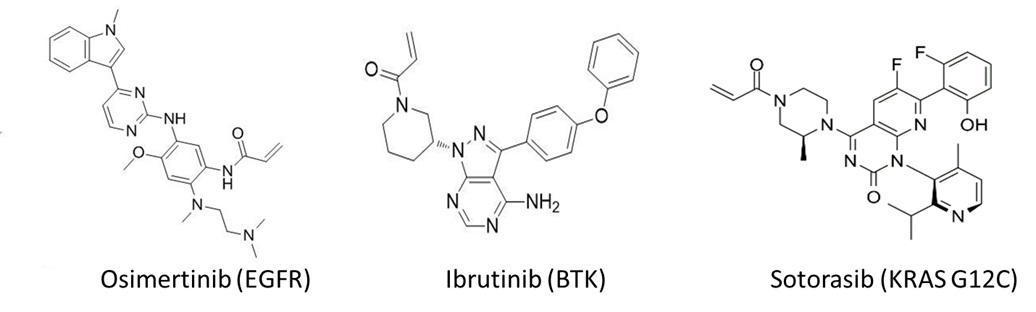
Figure 1. Examples of covalent drugs.
Direct screening for covalent libraries also provides an effective strategy for covalent ligand discovery, which is nicely exemplified by the discovery of KRAS G12C initial hits that led to the ultimate approval of Sotorasib (AMG510, Figure 1). Mass spectrometry, activity-based protein profiling (ABPP), as well as covalent DNA encoded libraries (DELs) represent the main experimental approaches. Although the first two approaches are straightforward and offer good selectivity, the throughput is relatively low, whereas the covalent DEL selection stands out with unique advantages of high throughput, low cost and fast investigation of large chemical space. On this front, HitGen has applied comprehensive exploration on covalent selection spanning DEL design, selection optimization, and compound characterization, leading to multiple successful covalent DEL screenings.
HitGen Covalent DELs
HitGen covalent DELs are composed of hundreds of millions of proprietary, drug-like binding motifs conjugated to tens of diverse covalent warheads (Figure 2, library design; Figure 3, examples of off-DNA structures). Specifically, binding motifs are constructed with several hundreds of proprietary scaffolds and more than 30,000 building blocks, resulting in over hundreds of millions of expandable compounds for covalent warheads to attach to. Different sets of covalent warheads are conjugated to these binding motifs based on the specific type of targets. For example, Michael acceptors and similar Cys-targeting electrophiles are used for cysteine-containing proteins. The expanded covalent warheads increase the probabilities of direct discovery of covalent inhibitors when the binding motifs are not strong enough, also the extended functionality on warheads may potentially pick up interactions therefore generating additional specificity. Similarly, the covalent DELs have specific sets of covalent warheads for Lys-, Ser- targeting proteins.
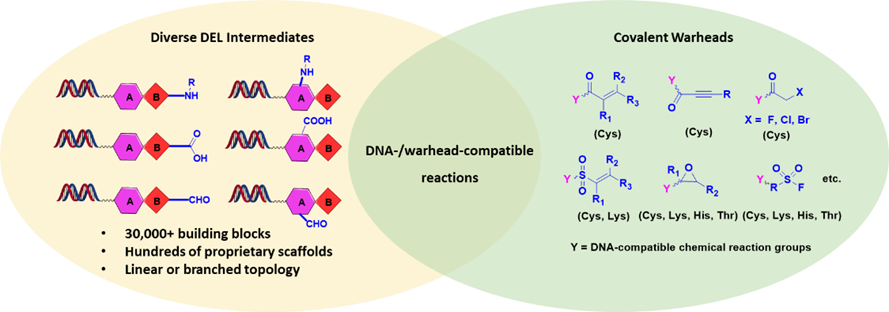
Figure 2. Covalent DEL design at HitGen.
At HitGen, the reactivities of the warheads are carefully examined and only the warheads with proper reactivity are chosen for covalent DEL synthesis, furthermore, the stabilities of these warheads are also carefully evaluated to ensure they are stable enough in the harsh conditions of DEL purification and DEL selection. The qualities of our covalent DELs, especially their warhead activities, are also periodically examined against representative targets.
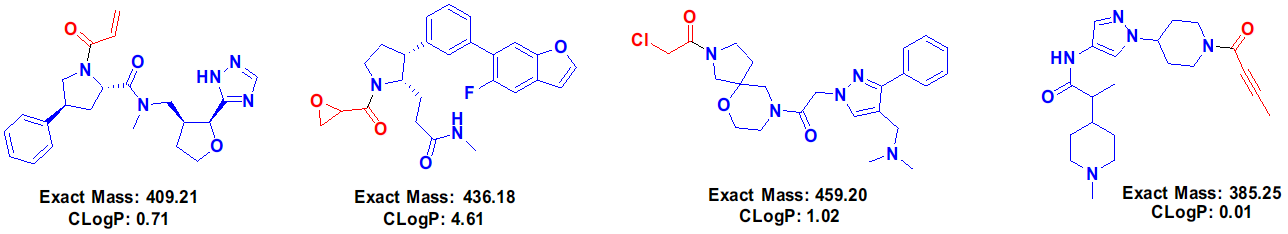
Figure 3. Examples of off-DNA structures of some HitGen covalent DEL compounds (warheads are shown in red).
Suitable Targets for Covalent Ligands
l Targeting kinases with non-conserved cysteine(s) near the ATP binding site to improve potency, and in some cases, to overcome drug resistance due to mutations (e.g. EGFR, BTK, JAK3)
l Targeting proteinases with crucial catalytic nucleophilic amino acid(s) (e.g. deubiquitinases, SARS-CoV-2 main proteases).
l Targeting enzymes with cysteine-ubiquitin conjugate formation during the catalysis (e.g. E2s, HECT and RBR E3s)
l Targeting other proteins with Cys, Ser, Lys, or Thr in or near the binding pocket for either inhibitor development or conversion to degraders.
Covalent DEL Selection
Given the “Bind and React” nature of covalent compounds, covalent DEL selections are performed with multiple parallel conditions to differentiate the compounds that bind and react with specific nucleophilic amino acid(s). Several DEL selection parameters including reducing agents, DEL input, DEL size and incubation time should be optimized before the selection. In addition, the positive control – DNA conjugate, mutants of nucleophilic residues and covalent tool compound blockings are frequently used to increase the confidence of finding positive hits. All these practices have been extensively explored at HitGen (Figure 4), leading to our comprehensive understanding of the covalent DEL selection process as well as demonstrated successful selection cases.
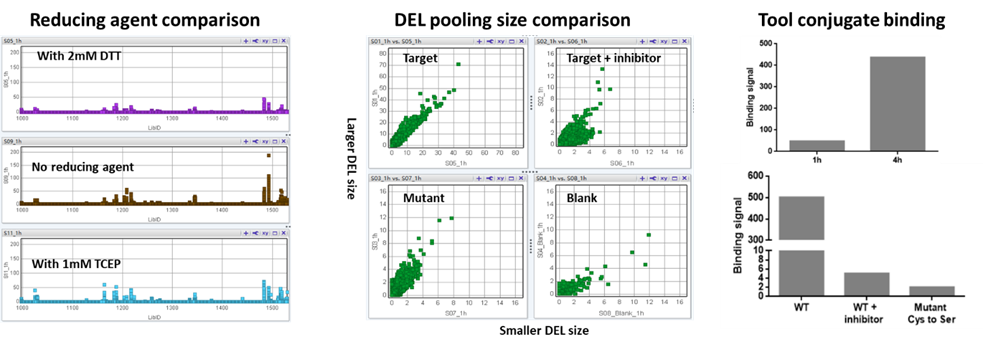
Figure 4. Covalent DEL selection exploration (selection of reducing agents, DEL pooling size comparison, selection of incubation times, inhibitor blocking and mutant comparison).
Case Study: identification of cysteine protease inhibitors
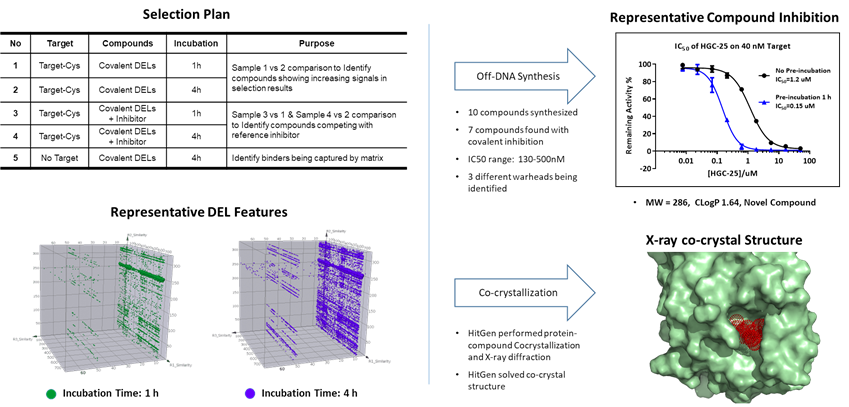
Figure 5. Covalent DEL selection plan, selection results, time-dependent inhibition and structure confirmation of covalent attachment to Cys.
Characterization of Covalent Ligands
Compared to classical reversible inhibitors, covalent compounds hold some unique features. First of all, the irreversibility reflects a time-dependent inhibition and a non-linear progression curve, compromising the accuracy of using IC50 for covalent compound profiling and ranking. Indeed, the Kinact/Ki represents a more meaningful parameter and has become the key metric for covalent compound characterization and the structure-activity-relationship (SAR) analysis. In addition, the irreversibility of the binding can also be validated by mass spectrometry, the jump-dilution experiment, dialysis, and the utilization of mutant proteins (Figure 6).
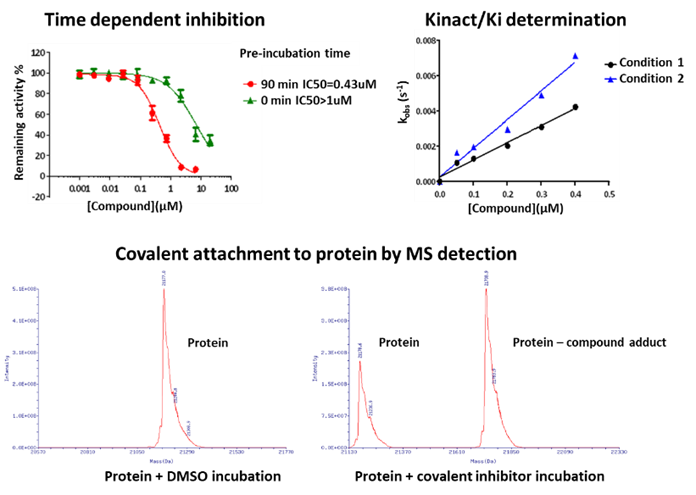
Figure 6. Representative covalent compound validation methods.
Outlook
Exploration of covalent ligands for direct inhibition of challenging targets has emerged as a very attractive approach for modern drug discovery. Covalent DEL screening, with its inherited large chemical space accessibility, less material consumption and the excellent screening efficiency, will be a very powerful approach to advance the field. With HitGen’s billion-scale, well-validated covalent DELs, rich experiences in DEL selection and comprehensive covalent profiling capabilities, your covalent ligand discovery effort will be significantly accelerated.



